HackTrack projects
The HackTrack is the official fun side of a Brainhack event, where people can work together on projects. What projects? Any kind! From exploding brains to resource gathering and data sharing!
Would you like to propose a project? Just open an issue on our GitHub repository and fill the template, we will be in touch to help you get going! But be sure to register first!
Do you plan to focus on visualization? Are you getting images so weird that they are kind of beautiful? Consider participating in the Beautiful Mistake category of the BrainArt SIG competition! You can find the submission form here.
Effect of Scan Quality on Pipeline Result Discrepancy
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
As we all know, neuroimaging experiment results are subject to reproducibility issues. Computing the same measure (on the same data) across multiple pipelines can yield different results. We will run FSL, FreeSurfer, and ASHS on a subset of Prevent-AD data to obtain volumetric measures across the three pipelines. We will then find the discrepancies in these measures between the pipelines, and see if we can explain the discrepancies given the properties of the input scans. We will do this by running MRIQC on the scans, and correlating the outputted image quality metrics with the pipelines discrepancies.
SKILLS
Python, FreeSurfer, FSL, ASHS

ChRIS Research Integration Service
github issue
Hubs: Glasgow, Americas
PROJECT DESCRIPTION
ChRIS is a platform for making reproducible, container-based analysis easy to develop and easy to use. Our goal for BrainHack is to get feedback on the application’s user experience, and do development work of wrapping existing software as ChRIS plugins.
SKILLS
Optional: docker, python, JSON
confounds: deconfounding library to properly handle confounds
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Develop a python library of methods to handle confounds in various neuroscientific analyses, esp. statistics and predictive modeling. More info and slides here: https://crossinvalidation.com/2020/03/04/conquering-confounds-and-covariates-in-machine-learning/
SKILLS
- python programming (preferably intermediate or better, but can work with basic skills)
- some statistics
- documentation ability

Create local (dev) installation package for brainlife.io
github issue
Hubs: Europe / Middle East / Africa, Glasgow
PROJECT DESCRIPTION
brainlife.io is a free and secure reproducible neuroscience analysis platform developed at the University of Texas and Indiana University and collaborators from around the world. It is funded by NSF/NIH. Although brainlife.io is SasS (software as a service) platform, we’d like to provide a capability to easily install brainlife.io on any VM on-prem. The initial goal of this is to allow external developers to more easily run/contribute to the platform development, but I’d like this capability to be used to install brainlife.io instances on other sites. We also need help to improve our documentation site (https://brainlife.io/docs) by identifying content that is confusing / lacking make any necessary content updates.
SKILLS
System Administration Docker bash/python scripting (Maybe High Performance Computing)
Creating the SPM M/EEG interface for Nipype
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
SPM is an open-source toolbox for analysing fMRI, M/EEG and even PET data, which had its first public release way back in 1991. SPM has been written (primary) in MATLAB, but Python users have been able to exploit a limited set of SPM’s functionality in their Python workflows via Nipype. We seek to widen the scope of what of SPM’s tools can be interfaced with in Nipype (and potentially its successor, Pydra) going forward. For BrainHack, our goal is to create the initial functionality to import, coregister, pre-process and source reconstruct MEG data, a modality currently not supported.
SKILLS
| Skill | level of expertise required |
|---|---|
| Python | beginner |
| SPM | confirmed |
| Git | 1 |
Creating a group-level modeling interface in AFNI using Python
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
There are quite a few powerful modeling tools in AFNI that perform group-level analysis. The programs range from simple to complex ones: GLM, ANCOVA with within-subject (or repeated-measures) factors, linear mixed-effects modeling, nonlinear modeling using smooth splines, intraclass correlation, Bayesian multi-level modeling, etc. These programs include 3dttest++, 3dMVM, 3dLMEr, 3dMSS, 3dISC, 3dICC, RBA, etc. Currently there is a Python program, gen_group_command.py, that provides a nice interface for the user to set up group-level analysis for GLM. Two potential projects are 1) create a unified user interface similar to gen_group_command.py that could generate most, if all, of the group-level programs; alternatively, 2) write a user interface for one or a few group-level programs.
SKILLS
Python Shell scripting
Physiopy - Documentation of Physiological Signal Acquisition Best Practices
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
Physiopy is a community formed around developing tools to operate physiological files in MRI setups. This project specifically will expand our package documentation containing tips and strategies on how to collect various forms of physiological data and the use of our packages. We aim to provide a succinct overview of what physiological data are typically recorded during an fMRI experiment, how these signals are recorded, and how these signals can improve our modeling of fMRI time series data. This is an active field of research, and we hope to encourage all users to get the latest recommendations prior to initiating a new study.
SKILLS
Excitement for learning the best practices in physiological signal acquisition. We welcome all contributions from any skill set and level, this project will predominantly be focused on documentation contributions. Git - we welcome any level of git (0-3), markdown knowledge helpful
ReproTube: YouTube for Reproducible Neuroimaging
github issue
Hubs: Glasgow, Americas
PROJECT DESCRIPTION
- establish automated workflow to update/maintain collection of videos, captions, metadata
- fix up automated captions
- push fixed up captions back to youtube (research feasibility for automation on that)
- use/work with DataCat (https://github.com/ohbm/hackathon2022/issues/53) project to provide a convenient front-end
- establish backup Some rough ideas for more “exciting” projects
- based on captions/metadata establish “recommendation engine”, express those in metadata, expose via DataCat
- establish automated extraction/annotation for extra features in videos, e.g.
- boundaries of sections
- slides/section titles
- extract acknowledgement/names from the slides (e.g. grant numbers etc) which otherwise would be missing from metadata/captions
- establish search engine to search up videos to answer a specific question, e.g.
- “run fmriprep on HPC”
- …. provide a query you would like to see …
- suggest more
SKILLS
We welcome all kinds of contributions from various skills at any level. It could range from proof-reading and fixing up captions, to setting up and writing documentation, discussing relevant functionality, or user-experience-testing, to bash and/or Python-based implementation (or refactoring) of the desired functionality and applying the infrastructure to other use cases, e.g. “MyLabTube” etc.
Implementation of test-retest reliability estimation in Python
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Accurately estimating reliability is crucial for many fields including neuroimaging and psychometrics. The traditional concept of intraclass correlation is not well-suited for task-related data at the trial level. Thus, a hierarchical modeling approach is desirable to more accurately capture the several levels involved in the data structure. A hierarchical modeling framework has already been implemented into the program TRR using R packages (e.g., brms) in AFNI. It would be a great addition to the neuroimaging and psychometrics communities if the modeling functionality is also implemented using Python. The project is looking for volunteers who are skilled in Python and familiar with some extent of hierarchical modeling. Useful links: https://github.com/adamhaber/pybrms
SKILLS
Python R
Implementation of region-based FMRI data analysis through hierarchical modeling using Python
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Hierarchical modeling is a powerful analytical framework that allows efficient information flow across multiple levels. Compared to the traditional mass univariate analysis, the hierarchical approach has the advantages of spatial specificity, high statistical efficiency, no discrimination against small regions, focus on effect estimation, full result reporting, etc. The modeling capability has already been implemented into the program RBA using R packages (e.g., brms) in AFNI. It would be a great addition to the neuroimaging community if the modeling functionality is also implemented using Python. The project is looking for volunteers who are skilled in Python and familiar with some extent of hierarchical modeling. Useful links: https://github.com/adamhaber/pybrms
SKILLS
Python R
Documentation and demo examples for BIDS Datasets: running FitLins with 3dREMLfit in AFNI
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
FitLins is the numerical engine developed to solve models for FMRI data analysis. 3dREMLfit is an AFNI program that fits subject-level time series data with the temporal correlations of the residuals characterized through an ARMA(1,1) structure. The output includes regression coefficients, their t- and F-statistics (plus the associated degrees of freedom). The functionality of 3dREMLfit has been incorporated into FitLins, but its usage has not been documented. This project solicits volunteers to write documents about model specification using FitLins with 3dREMLfit and to provide demonstrative examples. Below are some related links: https://github.com/nih-fmrif/code-convergence-summaries/issues/8 https://fitlins.readthedocs.io/en/latest/usage.html https://afni.nimh.nih.gov/pub/dist/doc/program_help/3dREMLfit.html
SKILLS
BIDS derivatives BIDS Stats Models Subject-level FMRI data analysis AFNI
BIDS Prov
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
Provenance is useful for interpreting and comparing scientific results as well as enabling reusable data and analysis. BIDS prov is a BIDS extension proposal to formalize a provenance framework for BIDS. The framework proposal is based on “w3c prov” and has been adapted for brain imaging. Currently the proposal is drafted in a google doc and contains most of the specifications needed but is lacking some illustrative examples. In particular, examples with BIDS-derivatives outputs using common neuroimaging software such as Freesurfer are challenging because not entirely formalized in the BIDS specification. The aim of this project is threefold:
- Provide complete examples of the BIDS-derivatives specification for
Freesurfer,SPM,FSL,AFNIor your favorite processing tool - Propose updates to the specs as needed
- Get community feedback on the overall proposal in order to submit a final version
SKILLS
Some experience with BIDS and/or neuroimaging software.

Bayes on the brain
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
To write a tutorial notebook for Bayesian approaches to analysing fMRI data. We aim to implement the equations from Friston & Penny (2007) Empirical Bayes and hierarchal models (SPM textbook). We also aim to simulate data and implement a basic analysis.
SKILLS
skills include going through text resources (SPM textbook), implementing equations/data simulation in python, writing plain language explanations of the equations, going through matlab/SPM functions for implementations, and reading/using the tutorial and providing feedback

The NARPS open pipelines project
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
NARPS is an fMRI many-analyst study in which 70 research teams were asked to analyze the same fMRI dataset with their favorite pipeline. We would like to take advantage of this wide range of pipelines to build a repository of existing fMRI pipelines that could be used to investigate the impact of analytical variability. Thus, we aim at reproducing these pipelines (for now, 8 were fully reproduced and validated) and to do so, we have access to a file containing textual descriptions of the pipelines as well as the original code for some of the pipelines. Our project can be split into several goals:
- Facilitate the access, the use and the contributions to the database with good documentation, the development of a standard way of contributing and the use of tools like Docker/Singularity to store data and directly execute the pipeline ;
- Decoding of the textual descriptions of the NARPS pipelines to understand what choices were made by researchers at each step of the analysis ;
- Code adaptation of each step using NiPype interface with the original software packages to build the workflow of the pipeline ;
- Computation of results and validation of the reproduction using original unthresholded and thresholded group level statistical maps published in NeuroVault ;
- Creation of a link with NeuroVault to directly publish the results on the platform.
SKILLS
We welcome all sorts of contributions. However, knowledge and practice in reproducibility and code sharing practices could help. One or more of the specific skills below would be greatly appreciated:
- Theoretical knowledge in fMRI data analysis pipelines
- Practical skills in fMRI data analysis pipelines with the software packages SPM, FSL and/or AFNI (others also appreciated)
- BIDS format
- Python programming language
- Experience with NiPype interface
- Docker/Singularity: installation and use of a Docker container, sharing images with Docker hub…
- Use of Git and Github
Expanding AFNI tutorials/examples
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
MRI processing can range from simple to complex. Having tutorials and demos helps people learn how to expand their processing repertoire, to make scripts more efficient, or to generate new ideas. AFNI contains a loooot of programs for a wide variety of tasks, across FMRI, DWI/DTI and structural MRI; many programs are even used for processing other modalities such as EEG, MEG, etc. Several tutorials and demos exist in the documentation: https://afni.nimh.nih.gov/pub/dist/doc/htmldoc/tutorials/main_toc.html …. but it would be great to add more. So, this project could create more tutorials/examples/demos, either with Python or shell scripting, and can even involve making a Sphinx-based RST page for adding to the website. Bring your own scripting ideas, and/or find out how to do processing with AFNI.
SKILLS
AFNI MRI processing interest/experience Sphinx/RST experience/interest shell scripting and/or Python
BIDS derivatives naming for afni_proc.py output
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
afni_proc.py creates a full pre-processing pipeline for a wide variety of MRI data, including alignment, motion correction, blurring, and regression modeling; outputs can be volumetric or projected onto surfaces, for single- or multi-echo data. Therefore, there is a variety of possible outputs. But afni_proc.py also has a dictionary of specific outputs that it populates, so the outputs are well-characterized and known. It would be nice to formulate rules for BIDS-derivative-appropriate naming for these outputs, for populating a new directory with these NIFTI, JSON and other files.
SKILLS
Some useful skills to have would be: BIDS derivatives experience FMRI processing interest/background AFNI Collaborative spirit! perhaps Python, if coding
Machine Learning for Stroke Lesion Segmentation
github issue
Hubs: Americas
PROJECT DESCRIPTION
We’d love for people interested in machine learning for image segmentation to try out our stroke lesion segmentation challenge! It’s built with the RAMP platform and meant to be a collaborative and educational way to get into new research areas. It has a starter kit for people to get familiar with some basic steps for creating a lesion segmentation algorithm, and learn about packages such as PyBIDS along the way. We just released it and hope people can find it useful to learn and work together on a complicated neuroimaging problem!
SKILLS
Familiarity with python/juptyer notebook

Review your tool with NMIND
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
NMIND is a collaborative initiative dedicated to responding to the growing frustration about redundancies in effort and reproducibility in neuroimaging. NMIND seeks to build a community which advances the development of standards in software development, nomenclature, and testing, to ultimately harmonize advancements in neuroscience. The goal of this project is to use the community-designed NMIND software-evaluation checklists to both a) evaluate a tool of interest, and b) feed-back the experience to improve the checklists themselves.
SKILLS
- None
Big Linear Modelling and Big Linear Mixed Modelling
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Large-scale, shared datasets are becoming increasingly commonplace in fMRI, challenging existing tools both in terms of overall scale and complexity of the study designs. As sample sizes grow, new opportunities arise to detect and account for grouping factors and covariance structures present in large experimental designs. To facilitate large sample analysis, we have created two Python toolboxes for use on HPC clusters:
- “Big” Linear Models (BLM); a toolbox for large-scale distributed fMRI Linear Model analyses.
- “Big” Linear Mixed Models (BLMM); a toolbox for large-scale distributed fMRI Linear Mixed Model analyses. At present, both tools are functioning and can be used for the analysis of tens of thousands of fMRI images. However, there is plenty that could be improved. Some of the goals we hope to address during the hackathon include:
- Developing a rigorous testing suite, potentially with continuous integration via Travis CI.
- Using Dask to streamline the current code base (at present there are a lot of bash scripts for ‘qsub’bing).
- Package releases. Neither of the toolboxes are currently on the Python Package Index.
- Adding customized covariance support. In previous work, we showed how the underlying methods BLMM uses could model custom covariance structures (e.g. AR, Diagonal, Toeplitz etc). However, at present BLMM does not support analyses with these features.
SKILLS
- Familiarity with the Python programming language (All goals).
- Familiarity with Travis CI (Goal 1 - recommended but not a necessity).
- Familiarity with Dask (Goal 2).
- Understanding of Linear Mixed Models and/or statistics (Goal 4).
Exploding Brains *in Julia*
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Particle simulations are used to generate visual effects (in movies, games etc…). In this project, I want to explore how we can use magnetic resonance imaging (MRI) data to generate interesting visual effects by using (2D) particle simulations. My aim is to convert my previous efforts (see 2020 OpenMR Benelux, 2020 OHBM Brainhack) to the Julia programming language. Why Julia? Because Julia offers convenient parallelization methods. This is very important for speeding up particle simulations, A compilation of my efforts so far can be seen at: https://youtu.be/_5ZDctWv5X4 I am planning to sophisticate the particle simulations and create new animations :)
SKILLS
| language | level of expertise required |
|---|---|
| Julia (necessary) | beginner |
| Python (would be useful) | confirmed |
| C++ (would be useful) | confirmed |
| Matlab (would be useful) | confirmed |

Neuroscout: A platform for fast and flexible re-analysis of (naturalistic) fMRI studies
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Neuroscout has two primary goals: 1) to democratize reproducible fMRI analysis by making it trivially easy to specify and fit models to public fMRI datasets and 2) facilitate the analysis of naturalistic datasets by leveraging machine-learning algorithms for automated annotation. Combined, these two goals seek to increase the reproducibility and generalizability of neuroimaging analysis. Neuroscout is already a stable platform. However, we are still working on growing our user base, and expanding the functionality. To that end we propose several goals for this hackathon: 1) Develop and get feedback on a end-to-end tutorial for Neuroscout. Currently, although Neuroscout features sufficient documentation, many users are not yet clear on how to best use the platform. By creating a complete end-to-end tutorial, and receiving feedback from the community, we aim to Neuroscout more accessible. This is a great issue for first time contributors! 2) Add more datasets. Neuroscout currently spans 40 distinct naturalistic stimuli across over a dozen independent datasets. However, many more datasets are made public yearly, and we will seek to further expand the number of datasets indexed. In particular, we hope to also include non-naturalistic datasets, to increase the scope of Neurocout. 3) Add more naturalistic features. Neuroscout uses machine-learning algorithms to annotate naturalistic stimuli such as movies. We developed a library (pliers) to provide a uniform API to diverse algorithms. However, there are many more algorithms that could be incorporated and tested. Many are available to extract but have not been actively worked on. A specific example would be to incorporate DeepGaze, to simulate eye tracking data in datasets without it. Validating new features by building models would be a relatively easy contribution for first timers. 4) Develop multivariate analysis pipelines. Neuroscout currently focuses on fitting multi-stage univariate GLM models using BIDS Stats Models (a formal specification). However, multivariate approaches are widely popular for analyzing naturalistic data. Although we don’t aim to fully specify multivariate models in a standardized format, we aim to prototype Neuroscout-compatible multivariate workflows that can take advantage of the vast number of datasets and features made easily available by the Neuroscout API. In the future, this project could be refined to become a core component of Neuroscout, as an alternative to GLM models.
SKILLS
Basic familiarity with neuroimaging analysis. Plus: tutorial writing skills, experience with naturalistic data and multivariate modeling (specifically encoding and decoding models).

Browbids - Make the browser run pybids
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Browsers (typically) don’t run Python code. That’s a shame, because the neuroimaging community has so many great tools written in Python that could be reused in interactive, graphical web apps without requiring users to install anything on their machine. If you’re working with BIDS, you are probably familiar with the bids-validator project that you can simply access with your web browser. Wouldn’t it be nice if you could just run some simple pyBIDS queries on your local BIDS dataset like that, directly in the browser, without having to first source your python environment and installing some libraries? And ideally without having to reimplement pyBIDS as a javascript library? Luckily, new solutions for python in the browser are currently being developed: Pyodide and pyscript are two (fairly) recent efforts to bring python to the browser using the webassembly language. These projects not only allow us to run Python code in the browser and use the output, but we can also install (some) Python libraries from PyPI and use them directly. So you can install ancp_bids inside pyodide, and then load a local BIDS dataset in your browser and run some basic queries. Here is a very simple prototype: https://browbids.netlify.app/ . But there are still a lot of challenges to solve, particularly with file system access and python dependencies that don’t play nice with pyodide. The rough goals for this hackathon are:
- understand what the most relevant pyBIDS use cases and queries are that can be implemented in the browser
- find out how we can make a reusable wrapper / plugin of the pyodide-pyBIDS bundle that other projects can just load to gain this functionality
- investigate ways to safely access the local filesystem so we can expose file metadata or even content to the python instance
- document what we have learned for other projects that may be also interested in browserizing a python library
- show a minimal but useful prototype of parsing a BIDS dataset in the browser using pyBIDS
SKILLS
We are very much starting from the beginning (although there is a simple proof of concept) and are trying to find a good way to address this project. Many of the initial challenges will probably require experience with javascript and web development as well as pyBIDS, but there is also a need for a BIDS user perspective to answer understand what would be useful things to do with pyBIDS in the browser. So any combination of
- experience using BIDS (to suggest or discuss use cases)
- good at writing accessible documentation / tutorials
- experience with Javascript (JS), any of
- ideally some accessible frontend framework (e.g. Vue)
- visualization libraries (e.g. d3)
- JS testing (e.g. Jest, cypress.io)
- packaging or distribution (e.g. npm / Vue plugin writing, …)
- browser filesystems, accessing local filesystem from browser,
- python
- familiarity with pybids / acnp_bids python API

Neurosynth-Compose User Testing
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
neurosynth-compose is a reimagining of the original neurosynth to give users the power to edit studies and create their own meta-analyses. While it is possible to create and execute a meta-analysis, we do not know what features/fixes people want. Goals:
- get people to test the platform and identify pain points
- identify places where documentation is unclear/missing
SKILLS
- interest in performing a meta-analysis
- patience to work with our website
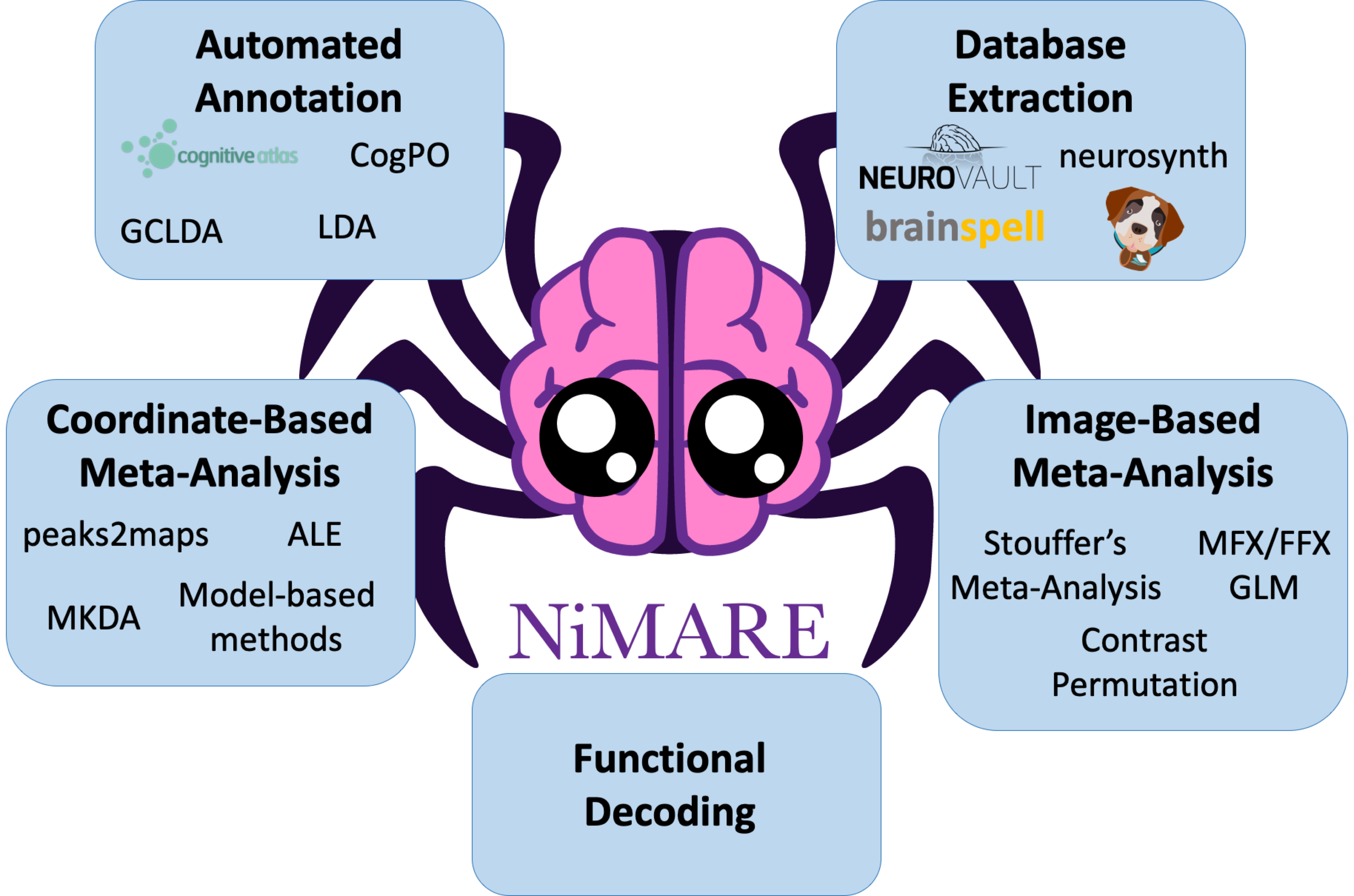
NiMARE: Neuroimaging Meta-Analytic Research Environment
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
User Guide Project
Currently, there is a mix of methods descriptions (the NiMARE Methods page) and examples of NiMARE’s functionality, but those two elements are very separate, and the examples are more a smattering of disconnected exhibitions than a tutorial. We came up with the idea of making the documentation more into a user guide.
Resources
- https://nimare.readthedocs.io/en/latest/methods.html
- https://nimare.readthedocs.io/en/latest/auto_examples/index.html
- https://github.com/neurostuff/ohbm2021-nimare-tutorial
- https://github.com/NBCLab/nimare-paper
User Guide Goals
- Create a jupyter book
- Leverage existing resources (see above)
- Write tutorial documentation with a novice meta-analysis practicitioner in mind
- Have people test jupyter book for typos/understanding/correctness
Extension addition project
NiMARE covers a wide breadth of meta-analytic algorithms. However, the only constant is change. With new algorithms emerging from research across the globe, there is a need for integrating the latest trends in neuroimaging meta-analysis and making the process smooth for new contributors. In order to kickstart the extension process the goals are twofold:
- integrate a new algorithm into NiMARE
- document the process and find pain points
SKILLS
Minimally, one of the following:
- meta-analysis: beginner
- python: beginner
Pydra: Converting existing scientific workflows to the new dataflow engine Pydra
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Pydra is a part of the second generation of the Nipype ecosystem. The goal of this project is to convert existing neuroimaging analytical workflows that are written in other languages to the new dataflow engine Pydra. The main deliverables of the project are comprehensive Pydra workflows that use interfaces from neuroimaging packages such as FSL, SPM, ANTs, AFNI, FreeSurfer. Goals for the Brainhack:
- Converting Nilearn.glm tutorial to Pydra
- Moving FitLins to Pydra
SKILLS
Minimal:
- Python
- Git Optional:
- Familiarity with GLM, BIDS
- Familiarity with Nipype, Nilearn

FLUX: A pipeline for MEG analysis and beyond
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
FLUX is a pipeline for analysing magnetoencephalography (MEG) data. By making the analyses steps and setting explicit, it aims to facilitate open science with the larger goal of improving the replicability of MEG research. So far, the FLUX pipeline has been developed for MNE-Python and FieldTrip with a focus on the MEGIN/Elekta system. The goal of this Brainhack project is to make the FLUX pipeline more flexible by making it fully BIDS compatible, as well as expanding its application to other systems, for instance CTF, optically pumped magnetometer (OPM) and electroencephalography (EEG).
SKILLS
This is an ongoing project and there are many ways in which you could contribute; from helping to improve the documentation to developing new functionalities, all kinds of contributions are welcome. Any of the following skills will be very helpful:
- experience with MEG or any other neurophysiological method
- basic Python/MATLAB knowledge
- familiarity with BIDS
- good writing skills
- being enthusiastic about Neuroscience!

Type hints for NiBabel
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Python has support for type annotations to help developers code more effectively by catching bugs via static analysis or making auto-complete suggestions. The more libraries that annotate their code with useful type hints, the more effective this assistance becomes. The goal of this project is to annotate NiBabel to ease the development process for neuroimaging in Python and improve the reliability of code built on top of NiBabel. We will use mypy for static analysis and test out type hinting in VScode.
SKILLS
Minimum
- Some experience with Python and numpy
Ideal
- Familiarity with some NiBabel APIs
- Experience with the typing and numpy.typing
DataCat: "bring your own data" and auto-generate user-friendly data catalogs
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
Summary
Do you want to learn how to generate a pretty and F.A.I.R. browser-based data catalog from metadata? Do you want to know how you can make your data known to the world, without sharing the actual data content on centralised infrastructure? Do you want to do this for free using open-source tools? YES?! Then “bring” your own data and join our hackathon project!
Overview
DataLad Catalog is a free and open source command line tool, with a Python API, that assists with the automatic generation of user-friendly, browser-based data catalogs from structured metadata. It is an extension to DataLad, and together with DataLad Metalad it brings distributed metadata handling, catalog generation, and maintenance into the hands of users. For a live example of a catalog that was generated using DataLad Catalog, see our StudyForrest Demo. The tool is now ready to be tested (and hopefully broken and then fixed!) on a wider range of user data. This is therefore intended to be a “bring your own data” project. If you are interested in metadata handling of (distributed) datasets, and specifically in generating a live catalog from said metadata, join us for a chance to turn your (metadata)data into a pretty browser application!
Project Goals
- Getting participants up to speed on what DataLad Catalog is and what it can do. This will be done through an initial discussion and by reading the primer
- Giving participants hand-on experience with the catalog generation process, with the use of walk-through tutorials
- Creating your own data catalogs
- Documenting feedback on your experience by creating issues (any and all types of issues are welcome!)
- Onboarding anyone interested in contributing to this tool in the many ways that are possible
SKILLS
We welcome all kinds of contributions from various skills at any level. From setting up and writing documentation, discussing relevant functionality, or user-experience-testing, to Python-based implementation of the desired functionality and creating real-world use cases and workflows. You can help us with any of the following skills:
- You have a dataset (or distributed datasets) for which you’d like to create an online catalog
- You enjoy breaking user interfaces or pointing out how the interface can be more intuitive
- You have experience with the Unix command line
- You are interested in creating accessible documentation
- You know Python / JavaScript / HTML / VueJS
- You are interested in learning about the DataLad ecosystem or the process of creating a DataLad extension
- You are interested in learning about the DataLad metadata handling capabilities and/or the process of creating a DataLad-based metadata extractors
- You have knowledge of metadata standards in your domain
- You have knowledge of BIDS and pybids (for the specific case of generating BIDS-related metadata, and rendering that in the catalog)
Nobrainer toolkit and model zoo for deep learning in neuroimaging
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
The goals of this hackathon are to improve usability of both the nobrainer library and the nobrainer model zoo.
SKILLS
- Python
Optional:
- Tensorflow
- Docker/Singularity

Exploring the AHEAD brains together
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
We recently made available a post-mortem data set including quantitative MRI and microscopy reconstructed in 3D at 200µm (see this article for details). The data set is openly accessible on FigShare already, but we would like to do more to help integrate it in other open science platforms to promote collaborative exploration of the data.
Goals for the Brainhack:
- set up a version of the data set that fits into Brainbox
- check how the data is handled by various visualization tools, make recommendations
- import initial parcellations from automated tools (nighres so far, but others could be run) into the visualizations
- manually annotate errors, artifacts, inconsistencies
- delineate new structures collaboratively
SKILLS
- enthusiasm for detail neuroanatomy
- dealing with data (re)formatting, header manipulation
- experience with various brain visualization tools
- ideas to increase collaboration in neuroanatomical atlasing

Brainhack Cloud
github issue
Hubs: Asia / Pacific, Glasgow, Europe / Middle East / Africa, Americas
PROJECT DESCRIPTION
A team of brainhack volunteers applied for Oracle Cloud Credits to support open source projects in and around brainhack with powerful cloud resources. This project is about learning about cloud computing and supporting other projects to make best use of the resources :)
SKILLS
Interest in cloud computing

Nilearn: Statistics for Neuroimaging in Python
github issue
Hubs: Europe / Middle East / Africa, Glasgow
PROJECT DESCRIPTION
Nilearn is an open-source Python package for fast and easy analysis and visualization of brain images. It provides statistical and machine-learning tools, with instructive documentation and a friendly community. It includes applications such as multi-voxel pattern analysis (MVPA), decoding, predictive modelling, functional connectivity, and brain parcellations. Moreover, in recent years, Nilearn has expanded to include Generalized Linear Models (GLMs) to analyse functional MRI data. For the Brainhack, we aim to get feedback from the community about some of the highlights of the latest release. These include:
- A new module nilearn.interfaces to implement loading and saving utilities with various interfaces
- Ability to provide custom hemodynamic response function (HRF) for dealing with non human primate data in GLM analysis
- Interactive surface plotting using Plotly engine
- Improved contributing documentation
Finally, we are always looking to get feedback on our documentation and to onboard new contributors.
SKILLS
We welcome all contributions from various skill sets and levels. This can include opening discussions around improvements to the documenation and/or code base, answering or commenting on questions or issues raised on github and neurostars, reviewing pull requests, and contributing code.

Higher order informational interactions in neuroimaging
github issue
Hubs: Europe / Middle East / Africa
PROJECT DESCRIPTION
Higher order interactions are being increasingly used and applied to neuroimaging data To date there isn’t a freely available python toolbox with proper input/output suitable for neuroimaging data. We have a pretty much optimized matlab code, and a functioning, yet not optimized python one by @PranavMahajan25 (with some improvements by @EtienneCmb). The goal(s) would be:
- improve the python implementation, adding statistical tests which are absent at the moment, adapting them from the matlab repository
- improve speed
- add input/output from BIDS processed data (MNE/NiLearn)
- explore solutions for plotting the data (some attempts by @renzocom) Discord server for the project
SKILLS
Python Matlab would help for the translation, but we can assist Some notions of statistics/probability theory could also help, but not necessary

watts_up_compute
github issue
Hubs: Glasgow
PROJECT DESCRIPTION
Integration of compute-tracker tools into neuroimaging pipelines to estimate carbon footprint of image processing. This is an ongoing project by the Sustainability and Environmental Action group (SEA-SIG) at the Organisation for Human Brain Mapping (OHBM). In this project we aim at better understanding the environmental costs of commonly used research pipelines and develop tools to help reduce them. Recently there have been several projects that track cpu/gpu “power draws” incurred during a compute task. These statistics can then be translated into carbon-footprint based on your location and time of processing. See these slides for more info!
Available trackers
Current implementations
- General purpose wrapper with CodeCarbon and EIT
- fMRIPrep integration with CodeCarbon
Brainhack tasks
- Test fMRIPrep integration on multiple hardware
- Integrate trackers into other neuroimaging pipelines e.g. FSL, SPM etc.
SKILLS
You don’t need to be familiar with all of these, just any subset of these would do!
Programming languages
- Python
- Bash
- Matlab
Neuro-software specific skills
- FreeSurfer
- fMRIPrep
- NiPype
- SPM
- FSL
Data standards
- Brain Imaging Data Structure (BIDS)
Git skills
- Git - 2: comfortable working with branches and can do a pull request on another repository

DataLad-Dataverse integration
github issue
Hubs: Glasgow, Europe / Middle East / Africa
PROJECT DESCRIPTION
Dataverse is open source research data repository software that is deployed all over the world in data or metadata repositories, so called Dataverse collections. It supports sharing, preserving, citing, exploring, and analyzing research data with descriptive metadata, and thus contributes greatly to open, reproducible, and FAIR science. DataLad, on the other hand, is a data management and data publication tool build on Git and git-annex. Its core data structure, DataLad datasets, can version control files of any size, and streamline data sharing, updating, and collaboration. In this hackathon project, we aim to make DataLad interoperable with Dataverse to support dataset transport from and to Dataverse instances. To this end, we will build a new DataLad extension datalad-dataverse, and would be delighted to welcome you onboard of the contributor team.
SKILLS
We plan to start from zero with this project, and welcome all kinds of contributions from various skills at any level. From setting up and writing documentation, discussing relevant functionality, or user-experience-testing, to Python-based implementation of the desired functionality and creating real-world use cases and workflows. You can help us with any of the following skills: You have used a Dataverse instance before and/or have access to one, or you are interested in using one in the future - You know technical details about Dataverse, such as its API, or would have fun finding out about them - You know Python - You have experience with the Unix command line - You are interested in creating accessible documentation - You are interested in learning about the DataLad ecosystem or the process of creating a DataLad extension - Your secret hobby is Git plumbing - You know git-annex, and/or about its backends - You want to help create metadata extractors for Dataverse to generate dataset metadata automatically
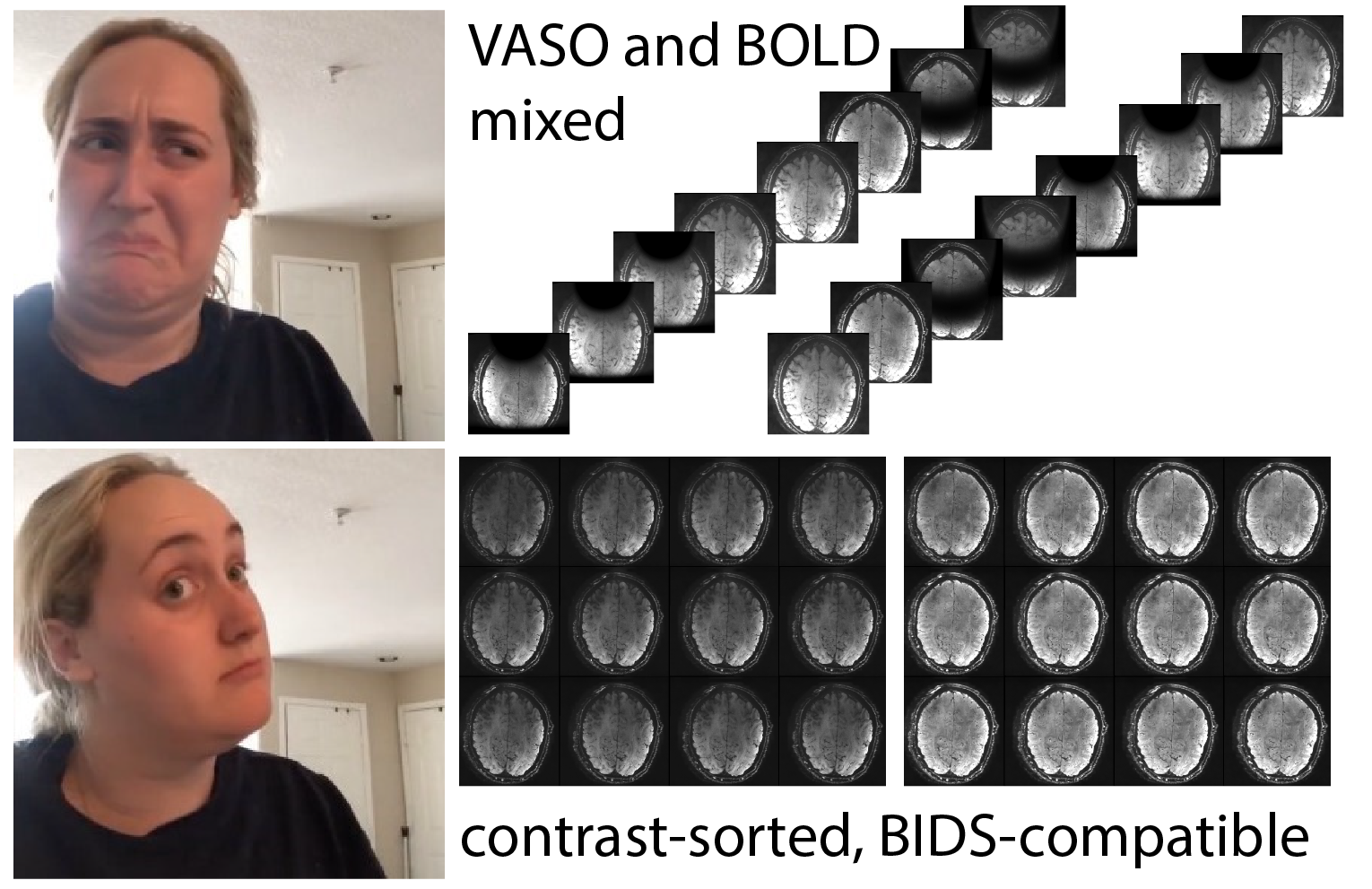
MOSAIC for VASO fMRI
github issue
Hubs: Europe / Middle East / Africa
PROJECT DESCRIPTION
Vascular Space Occupancy is an fMRI method that is popular for high-resolution layer-fMRI. Currently, the most popular sequence is the one by Rüdiger Stirnberg from the DZNE in Bonn, which is actively being employed at more than 30 sites. This sequence concomitantly acquires fMRI BOLD and blood volume signals. In the SIEMENS reconstruction pipeline, these signals are mixed together within the same time series, which challenges its user friendliness. Specifically:
- The “raw” dicom2nii-converted time-series are not BIDS compatible (see https://github.com/bids-standard/bids-specification/issues/1001).
- The order of odd and even BOLD and VASO image TRs is dependent on the nii-converter.
- Workarounds with 3D distortion correction, results in interpolation artifacts.
- Workarounds without MOSAIC decorators result in impracticable large data sizes.
The goal of this Hackathon is to extend the 3D-MOSAIC to solve these constraints. This functor is commonly used to sort images by echo-times, by RF-channels, by magnitude and phase in the SIEMENS reconstruction pipeline into sets of mosaics . However currently, this functor does not yet support the dimensionality of SETs. In this project we seek to include SETs into the capabilities of the functor.
Acknowledgements:
This project is based on previous tests by Rüdiger Stirnberg and Philipp Ehses to isolate the cause of the problem. This project will be based on the mosaic functor that was originally developed by Ben Poser and is currently being further extended by Philipp Ehses. The compatibility of the “raw” data with BIDS are supported by BIDS extensions spear-headed by Remi Gau and supported by Daniel Handwerker. The Hackathon logistics across various internet platforms are kindly guided by our Hackathon mentor Faruk Gulban.
SKILLS
- SIEMENS ICE programming in VE
- C++

NeuroCAUSAL - Development of an Open Source Platform for the Storage, Sharing, Synthesis and Meta-Analysis of Clinical Data
github issue
Hubs: Americas, Europe / Middle East / Africa
PROJECT DESCRIPTION
We wish to work with clinicians, neuroimagers, and software developers to develop an open source platform for the storage, sharing, synthesis and meta-analysis of human clinical data to the service of the clinical and cognitive neuroscience community so that the future of neuropsychology can be transdiagnostic, open, and FAIR.
Following the steps of what enable similar transition in functional neuroimaging, we are breaking down the over-ambitious goal in two stages:
- Create a sort of spin-off of Neuroquery that only covers lesion-related data hence allowing causal inferences
- A Neurovault kind of tool facilitating sharing of clinical data, which shall benefit from a sort of “neuropsyhcological BIDS formatting guidelines” (OHBM poster #2066 seems to have read our minds)
SKILLS
We are very heterogeneous in our own skills sets & levels and welcome all sorts of contributions 😄
The ontological issues we are facing require familiarity with neurology and/or cognitive science. These are contentious matters in the field and a perfect solution is not realistic: we seek the good compromise that will make this platform a useful tool for the broad community interested in the future of neuropsychology.
The technical issues will benefit from people familiar with tools to scrap data from texts, train/test predictive models, generally speaking converting to code our pipeline of papers selection > model fitting > function-to-structure mapping visualization (example).
We are aware we just started scratching the surface and will need lots of help on all fronts 🙏